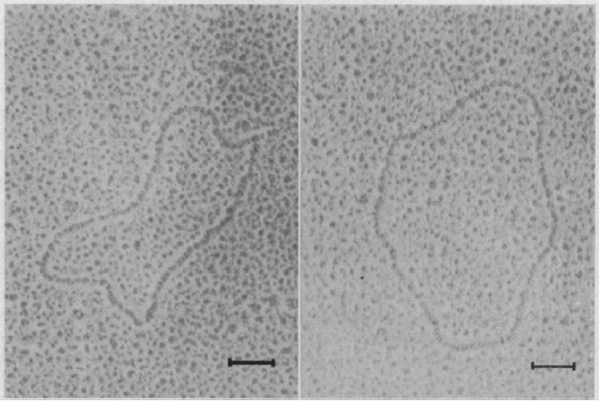
The main idea behind microarrays is that one nucleic acid (DNA) is immobilized on a solid support on a solid surface in a predefined location, and then another nucleic acid (RNA or a derivative such as fluorescently labeled complementary DNA) is hybridized to the surface. Microarrays were first developed in the 1990s by the laboratories of Patrick Brown at Stanford University and Jeffrey Trent, then at the National Institutes of Health (NIH). Beginning in the 1950s, Sol Spiegelman (1914–1983) pioneered the study of RNA hybridization to DNA (see ![]() http://profiles.nlm.nih.gov/PX/). By the early 1960s several groups had immobilized DNA on a solid support then hybridized purified RNA molecules under a variety of conditions. This figure shows electron micrographic images of circular DNA-RNA hybrids by Spiegelman and colleagues (Bassel et al., 1964). The bacteriophage
http://profiles.nlm.nih.gov/PX/). By the early 1960s several groups had immobilized DNA on a solid support then hybridized purified RNA molecules under a variety of conditions. This figure shows electron micrographic images of circular DNA-RNA hybrids by Spiegelman and colleagues (Bassel et al., 1964). The bacteriophage ![]() X174 was shown to transcribe RNA, which bound to DNA in a ribonuclease resistant complex. Studies such as these established the mechanisms by which DNA is transcribed to RNA, and ultimately led to the development of hybridization-based assays, including microarrays. The scale bar is 0.1 μm.
X174 was shown to transcribe RNA, which bound to DNA in a ribonuclease resistant complex. Studies such as these established the mechanisms by which DNA is transcribed to RNA, and ultimately led to the development of hybridization-based assays, including microarrays. The scale bar is 0.1 μm.
9
Gene Expression: Microarray Data ...
Get Bioinformatics and Functional Genomics, Second Edition now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

