Time machine for cancer diagnosis
Exciting new genetic testing technology has improved the speed and accuracy of cancer diagnosis.
 Cancer cells (source: The National Cancer Institute on Wikimedia Commons)
Cancer cells (source: The National Cancer Institute on Wikimedia Commons)
One of the most important improvements in oncology was the introduction of target therapy. It allowed clinicians to prescribe a pharmacological treatment specifically programmed to fight and kill only cancer cells, unlike wide-spectrum chemotherapy (Slamon D.J. et al. 2001).
To define a patient as eligible to receive target therapy, it is necessary to define the genetic profile of the cancer cells.
Among the techniques that have been developed in the past 20–30 years, one of the most used and considered the gold standard is in situ hybridization.
This technique is based on the principle of the specificity of the DNA sequences and uses genetic probes to recognize a specific gene, chromosome, or part of them.
This enables labs to see if there are numerical alterations, such as multiple copies of the genes; a reduction in the genes’ copies; or structural alterations such as deletions, inversions, or rearrangements; and then, to emit a report.
For example, in breast cancer it is important to define the number of the HER2 gene to assess the eligibility for the dedicated therapy; or in brain cancer where the loss of some parts of chromosome 1 and chromosome 19 is related to a specific cancer type (Slamon D.J. et al 2001; Barbashina V. et al 2005).
Today it is common to label these DNA probes with fluorescent dyes. Figure 1-1 shows one example of fluorescent in situ hybridization (FISH): the red and green dots are respectively two different genes. The blue big bodies are the cell nuclei.
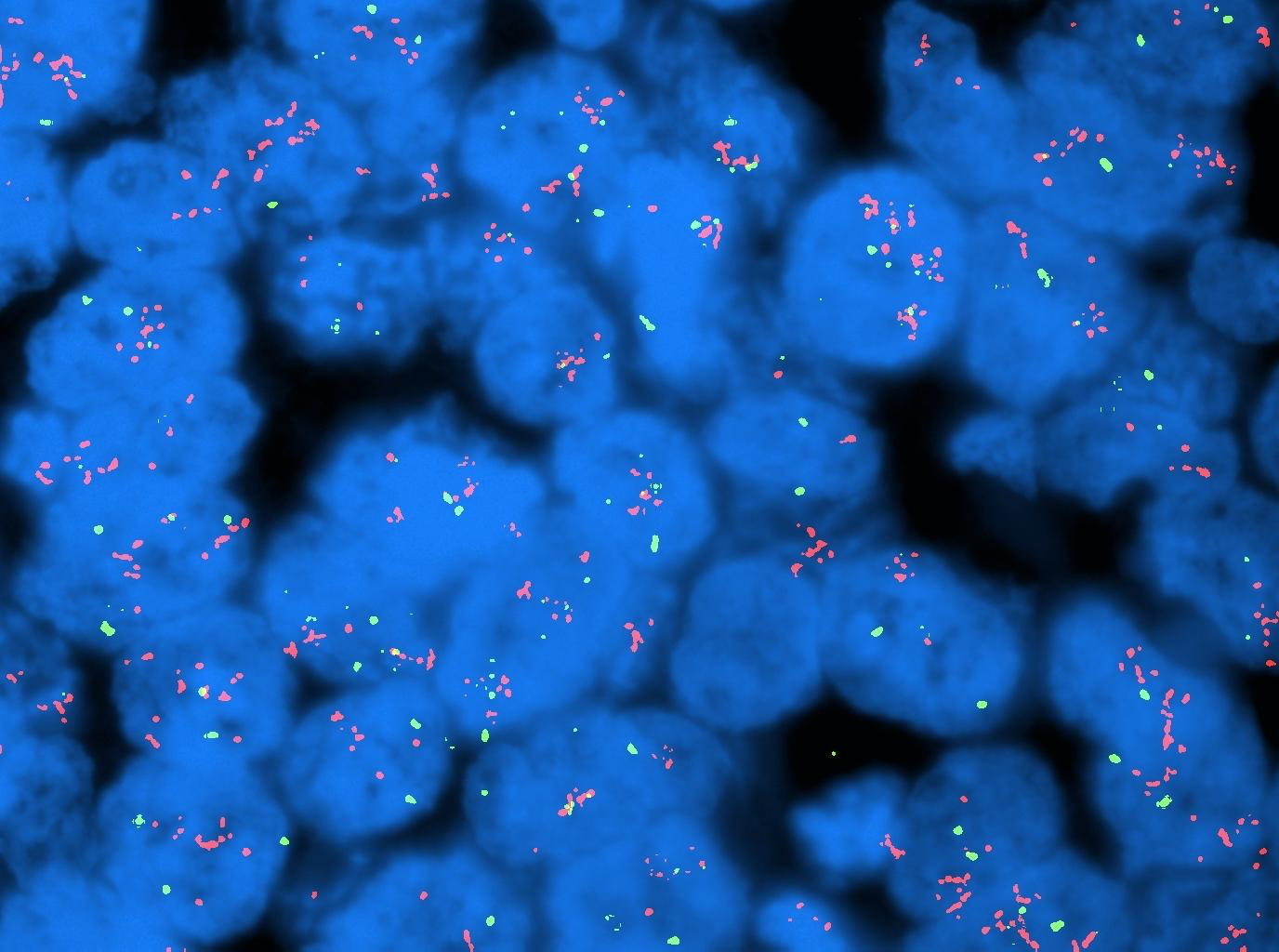
Following the last WHO guidelines for cancer diagnosis, the molecular characterization of an individual’s cancer cells is becoming mandatory, and it is fundamental to do a complete and accurate diagnosis.
The main problem is that these tests require at least two or three working days and are costly.
The wait reduces the lab’s capability to quickly emit a genetic report and also reduces the number of tests that could be performed daily.
The delay in diagnosis is not only a problem for the hospital administration in planning patients’ followup but is also related to an increasing amount of anxiety disorders among cancer patients (Baqutayan SMS 2012).
This limit of the FISH and the general ISH techniques started to become relevant once the new therapeutic targeted drugs for specific genetic assets were enlisted among the first-line treatments being prescribed to patients after their first biopsy.
Some examples of cancer treatment based on the ISH techniques results include the use of an antibody called Trastuzumab for the breast cancers with the amplification of the HER2 gene, or another antibody called Crizotinib, specific for a lung cancer type characterized by ALK gene genetic alterations (Voegel et al. 2002; Solomon B.J et al. 2014)
What Can Technology Do to Fix This Problem?
In the last 10 years, different approaches were used to reduce the time it takes to get a diagnosis and its cost.
One of them is a combination of engineering and biology: automate the diagnostic instead of performing it manually.
Automation will allow for tests to be run rapidly and with high accuracy. It will also reduce the cost of each test. However, there are still some limitations:
- The time required for a test procedure is about six to eight hours, even if automated, so it will still take two days to receive the report.
- The equipment is costly.
- Lab protocols will have to change to accommodate automation
- Labs will have to find a way to archive data properly such that the data are compatible with the readouts from the new equipment.
The other solutions proposed in the past four years are chemical-based (Dako-IQ FISH, and Abbott Intelli FISH, for example).
Most of them reduce the test time to at least four hours (or more) but have the same problems (e.g., high costs and incompatibility with lab reagents and procedures).
Is it possible to have a fast test without disrupting lab procedures?
Some second generations of these chemical solutions are available.
One of the solutions we developed, Rapid ISH integra, is on the market and allows labs to maintain their reagents while reducing the test time to only 2 hours and the cost by up to 50%.
The ISH techniques consist in a labeled DNA fragment that match its complementary fragment in the DNA of the sample. We can empower these techniques because our solution act by increasing the efficiency of the DNA interactions. To give you an idea let me make a parallelism among a DNA “meeting” and a meeting between friends:
The standard DNA-DNA hybridization act as the same as meeting a friend in a pub: you’re at the same table, but because of background noise, it’s harder to pay attention to each other.
With our products, we bring the meeting to a dedicated and isolated room in a library: no noise, no distraction, great efficiency.
Our reagents increase the efficiency of the DNA-DNA interactions while reducing the “noise” in the background and therefore the competition for the specific DNA regions in the sample making the test faster and more cost effective.
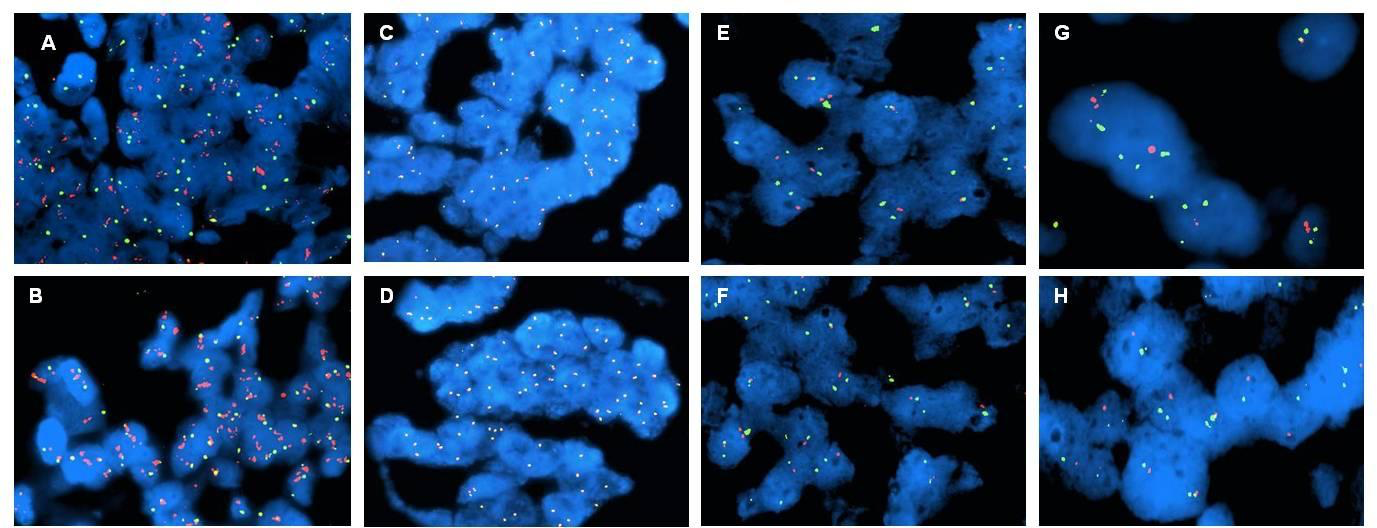
Figure 1-2 compares the standard FISH test and fast tests with Rapid ISH integra A-B HER2 gene (red dots) and chromosome 17 centromeric region (green dots) in breast cancer; C-D Alk gene (red dots) and eml4 gene (green dots) in lung cancer; E-F chromosome region 1p (red dots) and 1q (green dots) in brain cancer; G-H chromosome region 19q (red dots) and 19p (green dots) in brain cancer.
These markers are important both for the diagnosis and the therapy assignment.
In cancer diagnosis and research, the ISH techniques are not the only ones used but are part of an arsenal of techniques both DNA-based ( generically “genomics” ) and protein- based.
Thanks to the universal applicability on DNA of our chemical mixtures, we are working to improve the “genomics” so that labs can perform more tests more quickly.
We have also developed, thanks to participation in the RebelBio accelerator program, a new chemical-based technology that will improve the efficiency of the protein tests.
Currently the technology we developed is used both in human and in veterinary pathology to improve the efficiency of the ISH tests.
In the coming months we will start with global distribution due to the worldwide need for faster diagnostics and solutions.
Most of the technology, both in genomics and in informatics, is working to provide us quick and easy data and solutions.
We are working to make the technology better by combining our knowledge and passion and by cooperating with universities and companies.
We strongly believe in this cooperative approach to bring the science to the next level. Just as the ‘70s informatic revolution that gave us the personal computer, we want to make this technology accessible to all the cancer patients around the world.
Making the test more cost-effective allows labs in developing economies to adopt our technology and improve both the quality of care and opportunities to grow their scientific knowledge.
By making the test faster, we enable labs to increase their productivity (more tests run in parallel ) and their patients’ treatments and quality of life.
Selected References
- Slamon D.J. et al. 2001. “Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2.” N Engl J Med 2001; 344: 783-792. doi: 10.1056/NEJM200103153441101.
- Barbashina V. et al. 2005. “Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene.” Clin Cancer Res.11(3): 1119-28.
- Zneimer, Susan Mahler. 2016. Cytogenetic Laboratory Management: Chromosomal, FISH and Microarray-Based Best Practices and Procedures : Hoboken, New JerseyWiley-Blackwell.
- http://www.breastcancer.org/symptoms/testing/types/fish
- http://www.webmd.com/cancer/fish-cancer-test#1
- Baqutayan SMS. 2012. “The Effect of Anxiety on Breast Cancer Patients.” Indian Journal of Psychological Medicine.34 (2): 119–123. doi:10.4103/0253-7176.101774.
- https://www.cancer.gov/about-cancer/coping/feelings/stress-fact-sheet#q4
- https://www.cancer.org/treatment/treatments-and-side-effects/emotional-side-effects/anxiety-fear-depression.html
- Voegel et al. 2002. “Efficacy and Safety of Trastuzumab as a Single Agent in First-Line Treatment of HER2-Overexpressing Metastatic Breast Cancer.” Journal of Clinical Oncology. 20 (3): 719-726.
- Solomon B.J. 2014. “First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer.” N Engl J Med. 371: 2167–2177. doiI: 10.1056/NEJMoa1408440.
- http://bit.ly/2wPMzHM
- https://www.molecular.abbott/us/en/products/vysis-intellifish
- Muscatello L.V. Forthcoming. “Cat Loves Fish: HER2 Amplification Status in Feline Mammary Carcinoma.”
- Muscatello L.V. “HER2 Protein Overexpression and Gene Amplification in Feline Pulmonary Carcinoma.”
