Hidden Markov models
A Hidden Markov model, often abbreviated to HMM, which we will use here, is a Bayesian network with a repeating structure that is commonly used to model and predict sequences. In this section, we'll see two applications of this model: one to model DNA gene sequences, and another to model the sequences of letters that make up English text. The basic diagram for an HMM is shown here:
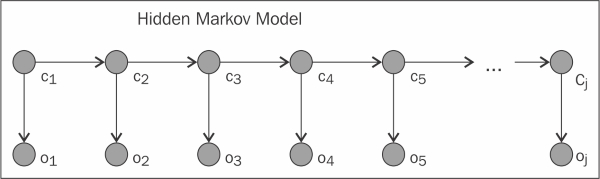
As we can see in the diagram, the sequence flows from left to right and we have a pair of nodes for every entry in the sequence that we are trying to model. Nodes labeled Ci are known as latent states, hidden states, or merely states, as they are typically ...
Get R: Predictive Analysis now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

