5.1 Motivation and Background
Random induced subgraphs arise in the context of molecular folding maps [24] where the neutral networks of molecular structures can be modeled as random induced subgraphs of n-cubes [17]. They also occur in the context of neutral evolution of populations (i.e., families of Qn2-vertices) consisting of erroneously replicating RNA strings. Here, one works in Qn4 since we have for RNA the nucleotides{A, U, G, C}. Random induced subgraphs of n-cubes have had an impact on the conceptual level [23] and led to experimental work identifying sequences that realize two distinct ribozymes [22]. A systematic computational analysis of neutral networks of molecular folding maps can be found in [11, 12]. An RNA structure, s, is a graph over [n] having vertex degree ≤ 1 and whose arcs are drawn in the upper half-plane (Figure 5.1). The set of s-compatible sequences, C[s], consists of all sequences that have at any two paired positions one of the 6 nucleotide pairs (A, U), (U, A), (G, U), (U, G), (G, C), (C, G). The structure s gives rise to a new adjacency relation within C[s]. Indeed, we can reorganize a sequence (x1,..., xn) into the tuple
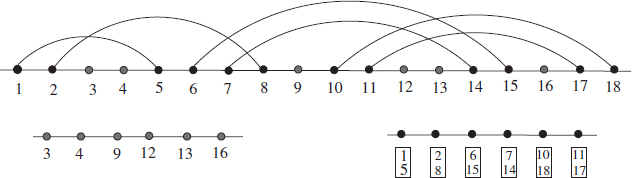
Figure 5.1 An RNA structure and its induced subcubes Qnu 4 and Qnp6: a structure allows one to rearrange its compatible sequences into unpaired and paired segments. The former is a sequence over the original alphabet A, U, G,
Get Analysis of Complex Networks now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

