Chapter 1. Introduction
We live in a time of great opportunity: technological advances are making it possible to generate incredibly detailed and comprehensive data about everything, from the sequence of our entire genomes to the patterns of gene expression of individual cells. Not only can we generate this type of data, we can generate a lot of it.
Over the past 10 years, we’ve seen a stunning growth in the amount of sequencing data produced worldwide, enabled by a huge reduction in cost of short read sequencing, a technology that we explore in Chapter 2 (Figure 1-1). Recently developed and up-and-coming technologies like long-read sequencing and single-cell transcriptomics promise a future filled with similar transformative drops in costs and greater access to ’omics experimental designs than ever before.

Figure 1-1. Recorded growth of sequencing datasets up to 2015 and projected growth for the next decade (top); growth in data production at the Broad Institute (bottom).1
The Promises and Challenges of Big Data in Biology and Life Sciences
Anyone, from individual labs to large-scale institutions, will soon be able to generate enormous amounts of data. As of this writing, projects that are considered large include whole genome sequences for hundreds of thousands of genomes. Over the next decade, we can expect to see projects sequencing millions of genomes and transcriptomes, complemented (and complicated) by a wide variety of new data types such as advanced cell imaging and proteomics. The promise is that the copious amounts of data and variety of new data types will allow researchers to get closer to answering some of the most difficult—if vexingly simple to ask—questions in biology. For example, how many cell types exist in the human body? What genetic variants cause disease? How do cancers arise, and can we predict them earlier? Because research is by its nature a team sport, we’ll want to broadly share much of the data on the horizon, we’ll want to share our algorithms to analyze this data, and we’ll want to share the findings with the wider world.
Infrastructure Challenges
The dual opportunities of falling costs and expanded experimental designs available to researchers come with their own set of challenges. It’s not easy being on the bleeding edge, and each new technology comes with its own complications. How do you accurately read single bases correctly as they whip on by through a nanopore? How do you image live cells in 3D without frying them? How do you compare one lab’s single-cell expression data with another lab’s while correcting for differences from batch effects? These are just a few examples in a very long list of technical challenges we face when developing or optimizing a new experimental design.
But the difficulty doesn’t stop with the data generation; if anything, that’s only the beginning. When the experiments are done and you have the data in hand, you have a whole new world of complexity to reckon with. Indeed, one of the most challenging aspects of ’omics research is determining how to deal with the data after it’s generated. When your imaging study produces a terabyte of data per experiment, where do you store the images to make them accessible? When your whole genome sequencing study produces a complex mixture of clinical and phenotypic data along with the sequence data, how do you organize this data to make it findable, both within your own group and to the wider research community when you publish it? When you need to update your methodology to use the latest version of the analysis software on more than 100,000 samples, how will you scale up your analysis? How can you ensure that your analytical techniques will work properly in different environments, across different platforms and organizations? And how can you make sure your methods can be reproduced by life scientists who have little to no formal training in computing?
In this book, we show you how to use the public cloud—computational services made available on demand over the internet—to address some of these fundamental infrastructure challenges. But first, let’s talk about why we think the cloud is a particularly appealing solution and identify some of the limitations that might apply.
This is not meant to be an exhaustive inventory of all available options; much like the landscape of experimental designs is highly varied, there are many ways to use the cloud in research. Instead, this book focuses on taking advantage of widely used tools and methods that include the Best Practice genomics workflows provided by the Genome Analysis Toolkit (GATK) (Figure 1-2), implemented using the Workflow Description Language (WDL), which can be run on any of the platform types commonly used in research computing. We show you how to use them in Google Cloud Platform (GCP), first through GCP’s own services, and then in the Terra platform operated on top of GCP by the Broad Institute and Verily.

Figure 1-2. GATK provides a series of Best Practices to process sequence data for a variety of experimental designs.
By starting with this end-to-end stack, you will acquire fundamental skills that will allow you to take advantage of the many other options out there for workflow languages, analytical tools, platforms, and the cloud.
Toward a Cloud-Based Ecosystem for Data Sharing and Analysis
From Figure 1-1, you can see that data has already been growing faster than the venerable Moore’s law can keep up, and as we discussed earlier, new experimental designs that generate massive amounts of data are popping up like mushrooms in the night. This deluge of data is in many ways the prime mover that is motivating the push to migrate scientific computing to the cloud. However, it’s important to understand that in its current form, the public cloud is mainly a collection of low-level infrastructure components, and for most purposes, it’s what we build on top of these components that will truly help researchers manage their work and answer the scientific questions they are investigating. This is all part of a larger shift that takes advantage of cloud-hosted data, compute, and portable algorithm implementations, along with platforms that make them easier to work with, standards for getting platforms to communicate with one another, and a set of conceptual principles that make science open to everyone.
Cloud-Hosted Data and Compute
The first giant challenge of this dawning big data era of biology lies in how we make these large datasets available to the research community. The traditional approach, depicted in Figure 1-3, involves centralized repositories from which interested researchers must download copies of the data to analyze on their institution’s local computational installations. However, this approach of “bringing the data to the people" is already quite wasteful (everyone pays to store copies of the same data) and cannot possibly scale in the face of the massive growth (both in dataset numbers and size) that we are expecting, which is measured in petabytes (PB; 1,000 terabytes).
In the next five years, for example, we estimate that the US National Institutes of Health (NIH) and other organizations will be hosting in excess of 50 PB of genomic data that needs to be accessible to the research community. There is simply going to be too much data for any single researcher to spend time downloading and too much for every research institution to host locally for their researchers. Likewise, the computational requirements for analyzing genomic, imaging, and other experimental designs is really significant. Not everyone has a compute cluster ready to go with thousands of CPUs at the ready.
The solution that has become obvious in recent years is to flip the script and “bring the people to the data.” Instead of depositing data into storage-only silos, we host it in widely accessible repositories that are directly connected to compute resources; thus, anyone with access can run analyses on the data where it resides, without transferring any of it, as depicted in Figure 1-3. Even though these requirements (wide access and compute colocated with storage) could be fulfilled through a variety of technological solutions, the most readily available is a public cloud infrastructure. We go into the details of what that entails in Chapter 3 as part of the technology primer; for now, simply imagine that a cloud is like any institution’s high-performance computing (HPC) installation except that it is generally a whole lot larger, a lot more flexible with respect to configuration options, and anyone can rent time on the equipment.
Popular choices for the cloud include Amazon Web Services (AWS), GCP, and Microsoft Azure. Each provides the basics of compute and storage but also more advanced services; for example, the Pipelines API on GCP, which we use when running analysis at scale in Chapter 10.

Figure 1-3. Inverting the model for data sharing.
Unlike the traditional HPC clusters for which you tend to script your analysis in a way that’s very much dependent on the environment, the model presented in Figure 1-3 really encourages thinking about the portability of analysis approaches. With multiple clouds vying for market share, each storing and providing access to multiple datasets, researchers are going to want to apply their algorithms to the data wherever it resides. Highly portable workflow languages that can run in different systems on different clouds have, therefore, become popular over the past several years, including WDL (which we use in this book and explore more in Chapter 8), Common Workflow Language (CWL), and Nextflow.
Platforms for Research in the Life Sciences
The downside of the move to the cloud is that it adds a whole new layer of complexity (or possibly several) to the already nontrivial world of research computing. Although some researchers might already have sufficient training or personal affinity to figure out how to use cloud services effectively in their work, they are undoubtedly the minority. The much larger majority of the biomedical research community is generally not equipped adequately to deal with the “bare” services provided by public cloud vendors, so there is a clear and urgent need for the development of platforms and interfaces tailored to the needs of researchers that abstract away the operational details and allow those researchers to focus on the science.
Several popular platforms present easy-to-use web interfaces focused on providing researchers a point-and-click means of utilizing cloud storage and compute. For example, Terra (which we explore in Chapter 11 and continue to use through the book), Seven Bridges, DNAnexus, and DNAstack all provide these sophisticated platforms to researchers over the web.
These and similar platforms can have different user interfaces and focus on different functionality, but at their core they provide a workspace environment to users. This is a place where researchers can bring together their data, metadata, and analytical workflows, sharing with their collaborators along the way. The workspace metaphor then allows researchers to run analysis—for example, on Terra this could be a batch workflow in WDL or Jupyter Notebook for interactive analysis—without ever having to dive into the underlying cloud details. We look at this in action in Chapters 11, 12, and 13. The takeaway is that these platforms allow researchers to take advantage of the cloud’s power and scale without having to deal with the underlying complexity.
Standardization and Reuse of Infrastructure
So it sounds like multiple clouds are available to researchers, multiple groups have built platforms on top of these clouds, and they all solve similar problems of colocating data and compute in places that researchers can easily access. The other side of this coin is that we need these distinct data repositories and platforms to be interoperable across organizations. Indeed, one of the big hopes of moving data and analysis to the cloud is that it will break down the traditional silos that have in the past made it difficult to collaborate and apply analyses across multiple datasets. Imagine being able to pull in petabytes of data into a single cross-cutting analysis without ever having to worry about where the files reside, how to transfer them, and how to store them. Now here’s some good news: that dream of a mechanism for federated data analysis is already a reality and is continuing to rapidly improve!
Key to this vision of using data regardless of platform and cloud are standards. Organizations such as the Global Alliance for Genomics and Health (GA4GH) have pioneered harmonizing the way platforms communicate with one another. These standards range from file formats like CRAMs, BAMs, and VCFs (which you will see used throughout this book), to application programming interfaces (APIs) that connect storage, compute, discovery, and user identity between platforms. It might seem boring or dry to talk about APIs and file formats, but the reality is that we want the cloud platforms to support common APIs to allow researchers to break down the barriers between cloud platforms and use data regardless of location.
Software architecture, vision sharing, and component reuse, in addition to standards, are other key drivers for interoperability. For the past few years, five US organizations involved in developing cloud infrastructure with the support of NIH agencies and programs have been collaborating to develop interoperable infrastructure components under the shared vision of a Data Biosphere. Technology leaders at the five partner organizations—Vanderbilt University in Nashville, TN; the University of California, Santa Cruz (UCSC); the University of Chicago; the Broad Institute; and Verily, an Alphabet company—articulated this shared vision of an open ecosystem in a blog post on Medium, published in October 2017. The Data Biosphere emphasizes four key pillars: it should be community driven, standards based, modular, and open source. Beyond the manifesto, which we do encourage you to read in full, the partners have integrated these principles into the components and services that each has been building and operating.
Taken together, community-based standards development in GA4GH and system architecture vision and software component sharing in Data Biosphere have moved us collectively forward. The result of these collaborative efforts is that today, you can log on to the Broad Institute’s Terra platform, quickly import data from multiple repositories hosted by the University of Chicago, the Broad Institute, and others into a private workspace on Terra, import a workflow from the Dockstore methods repository, and execute your analysis securely on Google Cloud with a few clicks, as illustrated in Figure 1-4.
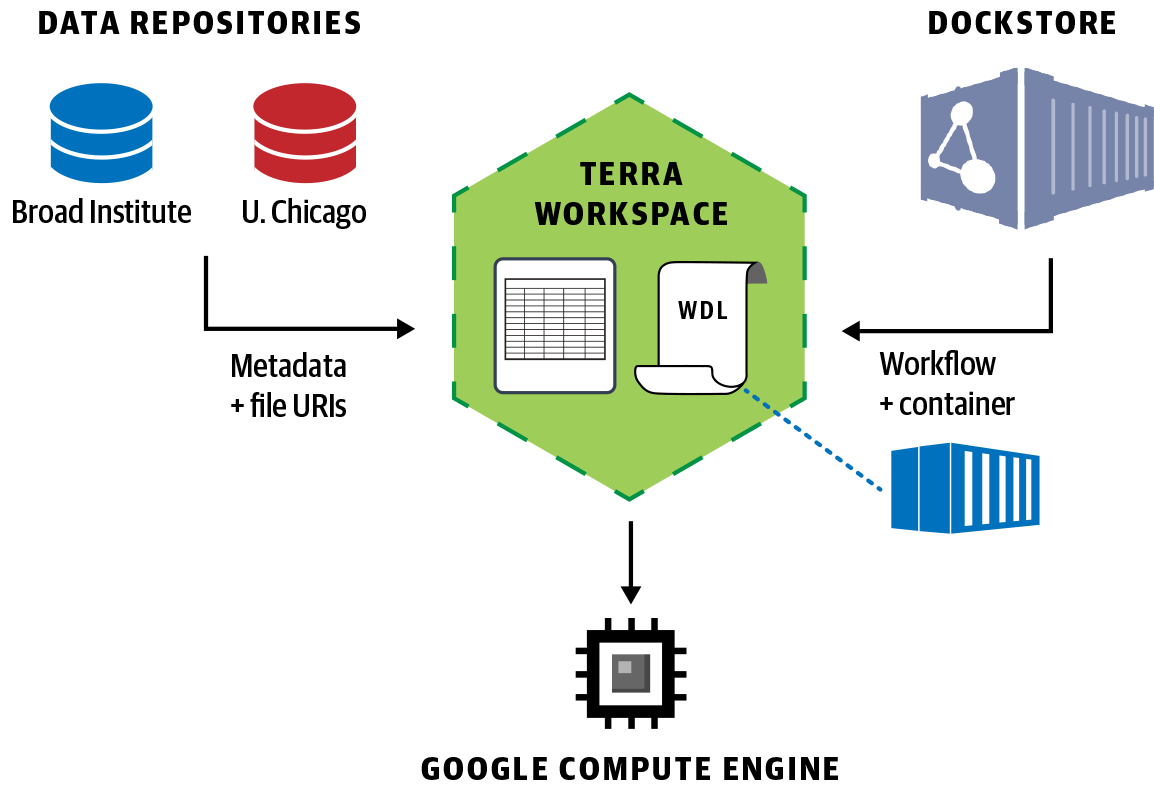
Figure 1-4. Data Biosphere principles in action: federated data analysis across multiple datasets in Terra using a workflow imported from Dockstore and executed in GCP.
To be clear, the full vision of a Data Biosphere ecosystem is far from realized. There are still major hurdles to overcome; some are, boringly, purely technical, but others are rooted in the practices and incentives that drive individuals, communities, and organizations. For example, there is an outstanding need for greater standardization in the way data properties are formally described in metadata, which affects searchability across datasets as well as the feasibility of federated data analysis. To put this in concrete terms, it’s much more difficult to apply a joint analysis to samples coming from different datasets if the equivalent data files are identified differently in the metadata—you begin to need to provide a “translation” of how the pieces of data match up to one another across datasets (input_bam in one, bam in another, aligned_reads in a third). To solve this, we need to have the relevant research communities come together to hash out common standards. Technology can then be used to enforce the chosen conventions, but someone (or ideally several someones) needs to step up and formulate those in the first place.
As another example of a human-driven rather than technology-driven stumbling block, biomedical research would clearly benefit from having mechanisms in place to run federated analyses seamlessly across infrastructure platforms; for example, cloud to cloud (Google Cloud and AWS), cloud to on-premises (Google Cloud and your institution’s local HPC cluster), and any multiplatform combination you can imagine on that theme. There is some technical complexity there, in particular around identity management and secure authentication, but one important obstacle is that this concept does not always align with the business model of commercial cloud vendors and software providers. More generally, many organizations need to be involved in developing and operating such an ecosystem, which brings a slew of complications ranging from the legal domain (data-use agreements, authority to operate, and privacy laws in various nations) to the technical (interoperability of infrastructure, data harmonization).
Nevertheless, significant progress has been made over the past several years, and we’re getting closer to the vision of the Data Biosphere. Many groups and organizations are actively cooperating on building interoperable cloud infrastructure components despite being in direct competition for various grant programs, which suggests that this vision has a vibrant future. The shared goal to build platforms that can exchange data and compute with one another—allowing researchers to find, mix, and match data across systems and compute in the environment of their choosing—is becoming a reality. Terra as a platform is at the forefront of this trend and is an integral part to providing access to a wide range of research datasets from projects at the NCI, National Human Genome Research Institute (NHGRI), National Heart, Lung, and Blood Institute (NHLBI), the Human Cell Atlas, and Project Baseline by Verily, to name just a few. This is possible because these projects are adopting GA4GH APIs and common architectural principles of the Data Biosphere, making them compatible with Terra and other platforms that embrace these standards and design philosophies.
Being FAIR
So far, we’ve covered a lot of ground in this chapter, starting with the phenomenal growth of data in the life sciences and how that’s stressing the older model of data download and pushing researchers toward a better model that uses the cloud for storage and compute. We also took a look at what the community is doing to standardize the way data and compute is made accessible on the cloud, and how the philosophy of the Data Biosphere is shaping the way platforms work together to make themselves accessible to researchers.
The benefits of this model are clear for platform builders who don’t want to reinvent the wheel and are motivated to reuse APIs, components, and architectural design wherever possible. But, from a researcher’s perspective, how do these standards from GA4GH and architecture from Data Biosphere translate to improvements in their research?
Taken together, these standards and architectural principles as applied in platforms like Terra enable researchers to make their research more FAIR: findable, accessible, interoperable, and reusable.2 We dive into this in more detail in Chapter 14. But for now, it’s useful to think that the work described up to this point by platform builders is all in an effort to make their systems, tools, and data more FAIR for researchers. Likewise, by embracing the cloud, writing portable workflows in languages like WDL, running analysis in Terra, and sharing workflows on Dockstore, researchers can make their own work more FAIR. This allows other researchers to find and access analytical techniques, interoperate, run the analysis in different places, and ultimately reuse tools as a stepping-stone to novel discovery. Throughout the book, we come back to the FAIR principles from the perspective of both platform builders and researchers.
Wrap-Up and Next Steps
Now that we’ve given you a background on some of the central motivations for why genomics as a discipline is moving to the cloud, let’s recapitulate how this book aims to help you get started in this brave new world, as outlined in the Preface. We designed it as a journey that takes you through a progression of technical topics, with an end goal of addressing the aforementioned infrastructure challenges, ultimately showing you how to get your work done on the cloud and make it FAIR to boot.
Remember, there are many different ways to approach these challenges, using different solutions, and we’re focusing on just one particular approach. Yet, we hope the following chapters will give you robust foundations to build on in your own work:
- Chapter 2 and Chapter 3
- We explore the fundamentals of biology and cloud computing.
- Chapter 5 through Chapter 7
- We dive into the GATK toolkit and the current Best Practices pipelines for germline and somatic variant discovery.
- Chapter 8 and Chapter 9
- We describe how to automate your analysis and make it portable with workflows written in WDL.
- Chapter 10 and Chapter 11
- We begin scaling up analysis first in Google Cloud, then in Terra.
- Chapter 12
- We complement workflow-based analysis with interactive analysis using Jupyter in Terra.
- Chapter 13 and Chapter 14
- We show you how to make your own workspaces in Terra and bring together everything you learned, to show you how to make a fully FAIR paper.
By the end of the book, we want you to have a good understanding of the current best practices for genomics data analysis, feel comfortable using WDL to express your analytical processes, be able to use Terra for both workflow-based and interactive analysis at scale, and share your work with your collaborators.
Let’s get started!
1 Stephens ZD, et al. “Big Data: Astronomical or Genomical?” PLoS Biol 13(7): e1002195 (2015). https://doi.org/10.1371/journal.pbio.1002195.
2 The FAIR Guiding Principles for scientific data management and stewardship by Mark D. Wilkinson et al. is the original publication of this set of principles.
Get Genomics in the Cloud now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

