Appendix F: Evolutionary Models of Molecular Phylogeny
CS Mukhopadhyay and RK Choudhary
School of Animal Biotechnology, GADVASU, Ludhiana
INTRODUCTION
Studies of molecular evolution and construction of phylogenetic trees are based on mathematical models that underpin the process of nucleotide substitution causing the process of speciation. A number of models have been hypothesized, based on certain assumptions of evolution. Some of the important models are described in this chapter.
Jukes–Cantor Model (1969) or JC69
This is the simplest DNA substitution model, which assumes equal base frequencies and a constant rate of evolution with a base substitution rate (Figure F1) of “α”. The transition (Purine to Purine and Pyrimidine to Pyrimidine) frequency is assumed to be same as the transversion (Purine to Pyrimidine and vice versa) frequency; hence, the model has a single parameter “α”. A substitution matrix (Figure F2) is generated for all possible base substitutions, assuming a fixed rate of changeover.
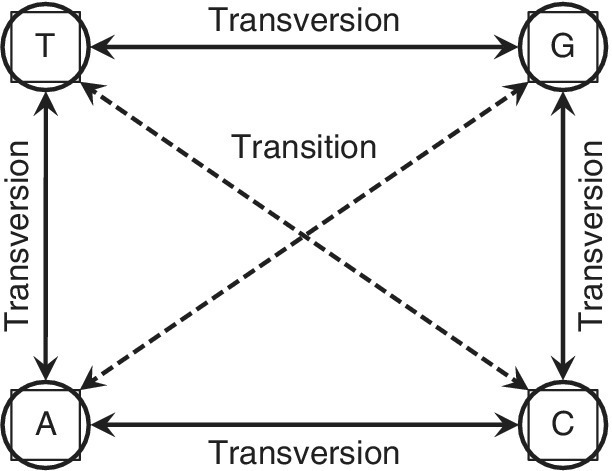
FIGURE F1 Substitution of nucleotides leading to transition and transversion.
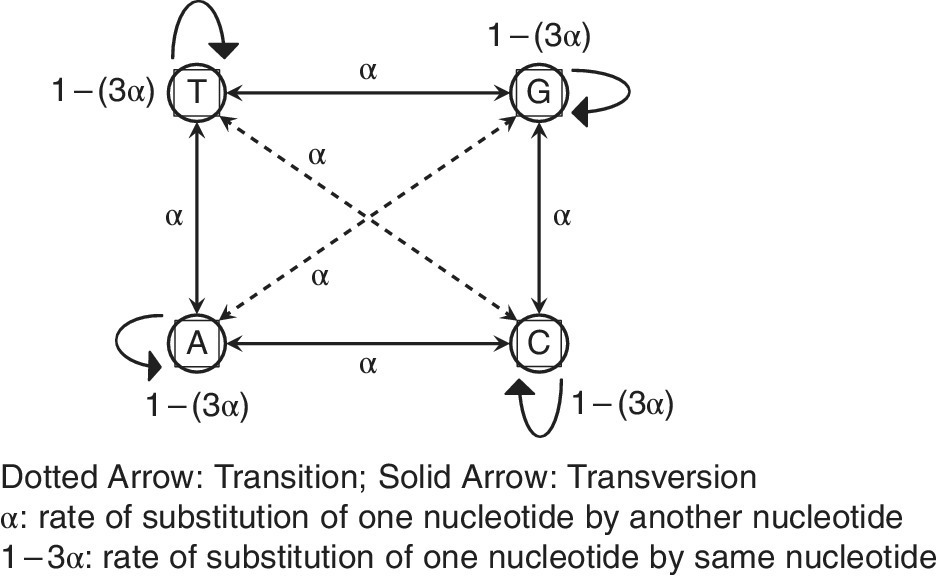
FIGURE F2 Jukes–Cantor one‐parameter substitution model
(Jukes and Cantor, 1969).
The substitution matrix is generated from a constant rate of base substitution per unit time, based on the JC69 ...
Get Basic Applied Bioinformatics now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

